1. What is RCDdb?
Regulated cell death (RCD) is a special form of regulated cell death caused by the activation of one or more signal transduction modules, which is important for maintaining biological balance and intracellular homeostasis. In recent years, the research on cell death has gradually increased, and the genes related to different cell death types have been accumulated in the database. For example, FerrDb, HAMdb and HADb databases provide genes related to ferroptosis and autophagy. Existing studies have shown that the molecular mechanisms of different RCD modes exhibit a considerable interconnection and synergy. XDeathDB provides molecular interactions of different cell death types and annotation information. However, there are several deficiencies in the current data resources. First, most databases only support a certain type of death data, which is not conducive to large-scale discovery and association research. Second, there are fewer numbers of genes and types of cell death such as ‘Disulfidptosis’ not be covered. Third, most of them don’t support disease association analysis and data visualization. Therefore, it is highly desirable to construct a comprehensive RCD resource and interactive analysis platform, which will benefit researchers in the field to study and explore cell death.
2. Database Content and Construction
Here, we established a comprehensive manually curated RCD database (chenyclab.com/RCDdb/) for RCD related gene sets and the detailed annotation information. Through collection and screening 2174 published literatures and searching according to key words, RCDdb has recorded 10 known and 5 newly reported death types and a total of 1849 RCD genes and evidence description of the experiment, including 1603 coding genes and 246 non-coding genes. RCDdb provides 49 visualizations, RCDdb also provides annotation of different types of RCD genes, including disease, drug, pathway, clinical, mutation, expression and interaction network. RCDdb provides a user-friendly interface to search, browse, and visualize detailed information on different RCD genes. In addition, RCDdb provides three RCD-based online analysis tools: 1) Enrichment analysis of user's interested genes to the RCDdb reference gene sets. 2) Screening signature RCD genes by three machine learning algorithms. 3) Gene set scoring analysis by GSVA and ssGSEA method. RCDdb is a powerful platform that provides a variety of types of RCD gene sets, annotations and analysis based RCD gene sets for users.
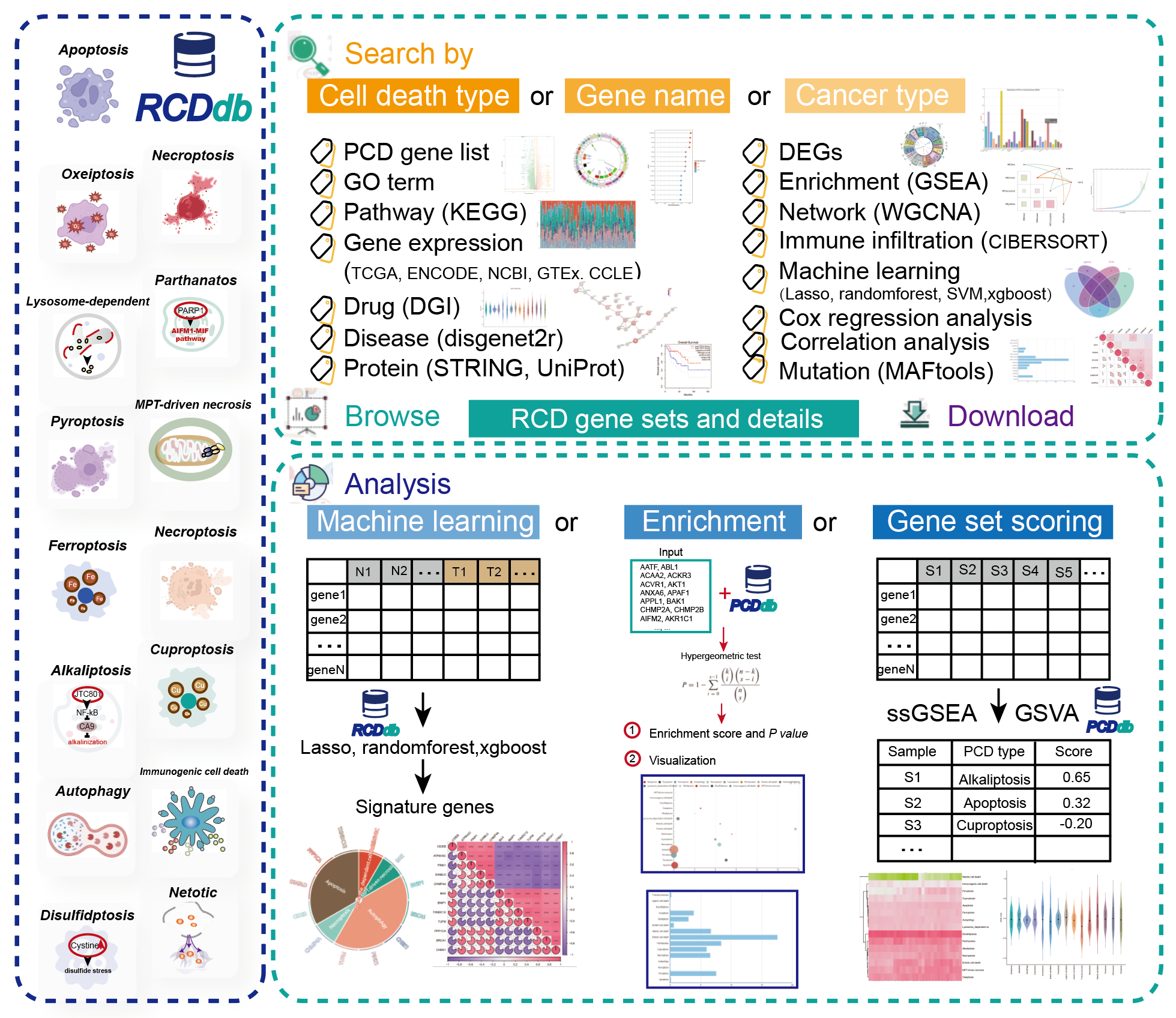

3.How to Use the RCDdb?
The browsing page is organized into an interactive table that allows users to quickly search for RCD genes and customize filters based on RCD type. On the left RCDdb also provide a timeline of 15 RCD discoveries. Users can click the "Show entries" drop-down menu to change the number of records displayed per page. When users select a RCD type, the right main table will show the RCD type, gene name, gene description, gene type, PMID and related comments. The browse tables are available for download.




RCDdb provides three query methods, including RCD type-based, RCD gene-based, and cancer-based queries.
Search RCD genes by death type. Users can select a RCD type and the return page will contain seven sections corresponding to the RCD type. The overview section presents basic information about this RCD type. The GO and KEGG annotation part includes 14146 GO pathways and 1090 KEGG pathways with a p-value less than 0.05 obtained by clusterProfiler analysis, and is displayed by histogram and line graph.In the disease annotation part, we used disgenet2r to analyze the data of RCD gene-disease association confirmed by literature reports in the artificially curated CURATED database, a total of 69536 records. In the drug annotation section, we downloaded all gene-drug interaction information from the DGI database, and retained a total of 17,690 gene-drug interaction information after screening RCD genes. In the protein annotation section, we downloaded the STRING database v11.5 version of the dataset and screened and retained the protein annotations of 78,814 RCD genes. And we also collected 3088 UNIPORT database annotations of all RCD genes. In the transcription and expression part, we collected 7 different types of transcriptome data from TCGA, ENCODE, NCBI, GTEx, and CCLE databases, analyzed the FPKM value of RCD genes, and displayed them in a heat map. Users can click the button to view any Transcriptome expression levels.

Search RCD gene by gene name. Users can enter the name of the gene of interest to obtain the annotation information of the RCD gene, and the returned page contains 6 parts. In the gene information section, the basic information of the gene is displayed, including SYMBOL description, gene type, Ensembl ID, which type of death it belongs to, as well as supporting literature sources and corresponding annotations. In addition, we provide links to 8 mainstream databases, and users can click on the icon links to view more abundant gene information. In the Diseases, Drugs, and Annotations section, we added these annotation information and a gene interaction network diagram. In the protein annotation section, we also provide the 3D protein structure predicted by AlphaFold for display. In the survival analysis section, users can freely choose genes and any of the 33 types of cancer for survival analysis, and provide users with a wealth of options for personalized analysis. In the transcription expression section, the transcription expression level of the gene in different samples is displayed in the form of a histogram.


Search RCD gene by cancer type. There is a close relationship between programmed cell death and cancer. In the development and progression of cancer, the uncontrolled and abnormal programmed cell death is often one of the key biological events. Therefore, we analyzed the genomes, transcriptional Group, proteome and corresponding clinical information data, and extracted the key RCD genes, so that biologists can easily explore the connection between RCD and cancer. Users can choose any type of cancer to search, and the result page provides 20 kinds of analysis, and rich visual chart display of the results. We analyzed the differential expression of RCD genes in this cancer and the enrichment and annotation of GO and KEGG, GSEA, WGCNA analysis, PPI network, tumor signal analysis and immune infiltration analysis. In particular, we extracted characteristic genes for all differential RCD genes through four classical machine learning algorithms, and performed correlation analysis, COX regression analysis, genomic mutation analysis and survival analysis on these characteristic genes in tumor samples to explore the effect of these characteristic genes on effect on tumorigenesis.


Search RCD genes by death type. Users can select a RCD type and the return page will contain seven sections corresponding to the RCD type. The overview section presents basic information about this RCD type. The GO and KEGG annotation part includes 14146 GO pathways and 1090 KEGG pathways with a p-value less than 0.05 obtained by clusterProfiler analysis, and is displayed by histogram and line graph.In the disease annotation part, we used disgenet2r to analyze the data of RCD gene-disease association confirmed by literature reports in the artificially curated CURATED database, a total of 69536 records. In the drug annotation section, we downloaded all gene-drug interaction information from the DGI database, and retained a total of 17,690 gene-drug interaction information after screening RCD genes. In the protein annotation section, we downloaded the STRING database v11.5 version of the dataset and screened and retained the protein annotations of 78,814 RCD genes. And we also collected 3088 UNIPORT database annotations of all RCD genes. In the transcription and expression part, we collected 7 different types of transcriptome data from TCGA, ENCODE, NCBI, GTEx, and CCLE databases, analyzed the FPKM value of RCD genes, and displayed them in a heat map. Users can click the button to view any Transcriptome expression levels.

Search RCD gene by gene name. Users can enter the name of the gene of interest to obtain the annotation information of the RCD gene, and the returned page contains 6 parts. In the gene information section, the basic information of the gene is displayed, including SYMBOL description, gene type, Ensembl ID, which type of death it belongs to, as well as supporting literature sources and corresponding annotations. In addition, we provide links to 8 mainstream databases, and users can click on the icon links to view more abundant gene information. In the Diseases, Drugs, and Annotations section, we added these annotation information and a gene interaction network diagram. In the protein annotation section, we also provide the 3D protein structure predicted by AlphaFold for display. In the survival analysis section, users can freely choose genes and any of the 33 types of cancer for survival analysis, and provide users with a wealth of options for personalized analysis. In the transcription expression section, the transcription expression level of the gene in different samples is displayed in the form of a histogram.


Search RCD gene by cancer type. There is a close relationship between programmed cell death and cancer. In the development and progression of cancer, the uncontrolled and abnormal programmed cell death is often one of the key biological events. Therefore, we analyzed the genomes, transcriptional Group, proteome and corresponding clinical information data, and extracted the key RCD genes, so that biologists can easily explore the connection between RCD and cancer. Users can choose any type of cancer to search, and the result page provides 20 kinds of analysis, and rich visual chart display of the results. We analyzed the differential expression of RCD genes in this cancer and the enrichment and annotation of GO and KEGG, GSEA, WGCNA analysis, PPI network, tumor signal analysis and immune infiltration analysis. In particular, we extracted characteristic genes for all differential RCD genes through four classical machine learning algorithms, and performed correlation analysis, COX regression analysis, genomic mutation analysis and survival analysis on these characteristic genes in tumor samples to explore the effect of these characteristic genes on effect on tumorigenesis.


RCD gene set enrichment analysis to predict the type of cell death associated with the gene set of interest to the user. The user inputs a list of genes of interest or a text file containing genes of interest, and the RCDdb maps these genes into the reference RCD gene sets and uses hypergeometric tests to calculate statistical significance for enrichment and similarity scores. The return page displays the enrichment analysis results. Users can click the green button at the top to download the result table and draw the enrichment analysis bubble chart and bar chart. Each column in the table represents the RCD type, the number of RCD genes, proportion, Jaccard score, Simpson score, enrichment analysis P-value and adjusted P-value. Users can click the small icon on the far right to view the detailed information of the set and similar sets. Users can also click on the count column hyperlink to get the names of genes tagged to the collection and perform survival analysis on them.
 RCD signature gene analysis. RCDdb provides a lasso, randomforst, xgboost three classic machine learning algorithms to screening of RCD signature genes. The user needs to input a normalized expression matrix and the sample group information. Then RCDdb will extract RCD feature genes in the matrix according to the corresponding machine learning algorithm. The left table of the result page shows the feature genes and their corresponding RCD type, and the sunburst chart is visualized on the right. The middle two plots show the training model information, and the bottom two plots show the expression correlation results between the signature genes.
RCD signature gene analysis. RCDdb provides a lasso, randomforst, xgboost three classic machine learning algorithms to screening of RCD signature genes. The user needs to input a normalized expression matrix and the sample group information. Then RCDdb will extract RCD feature genes in the matrix according to the corresponding machine learning algorithm. The left table of the result page shows the feature genes and their corresponding RCD type, and the sunburst chart is visualized on the right. The middle two plots show the training model information, and the bottom two plots show the expression correlation results between the signature genes.
 RCD gene set scoring analysis to calculate the RCD score for each sample of the expression matrix. First, users need to enter a normalized expression matrix. RCDdb provides two scoring algorithms, GSVA and ssGSEA to choose for users. Then RCDdb will perform RCD gene set scoring analysis on the input expression matrix. The result table is displayed, and each column in the table is the sample information, RCD type, and corresponding RCD score. The heatmap visualizes the scores of each sample in each RCD type, and the violin chart visualizes the high and low scores of each RCD type in all samples.
RCD gene set scoring analysis to calculate the RCD score for each sample of the expression matrix. First, users need to enter a normalized expression matrix. RCDdb provides two scoring algorithms, GSVA and ssGSEA to choose for users. Then RCDdb will perform RCD gene set scoring analysis on the input expression matrix. The result table is displayed, and each column in the table is the sample information, RCD type, and corresponding RCD score. The heatmap visualizes the scores of each sample in each RCD type, and the violin chart visualizes the high and low scores of each RCD type in all samples.




All gene sets of RCD type have been arranged and sorted into separate files for download in our database. RCDdb also provide two types of file formats for download, including ‘.csv’ and ‘.txt’. Users can download our reference collection as a data supplement and positive collection for customized research.


4. Frequently Asked Questions
4.1 Why might web pages load slowly?
Reply: RCDdb has advanced storage technology and sufficient bandwidth to meet the needs of most users for the speed of web page loading. However, it is not excluded that few users have poor user experience due to network reasons.4.2 What is the difference between the signature genes and all genes?
Reply: The signature gene refers to the gene with significant characteristic expression in two modes (eg:tumor and normal).
5.Development Environment
The current version of RCDdb was developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). PHP 7.0 (http://www.php.net) was used for server-side scripting. The interactive interface was designed and built using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). ECharts (https://www.echartsjs.com/) and Highcharts (https://www.highcharts.com.cn/) were used as a graphical visualization framework. We recommend to use a modern web browser that supports the HTML5 standard, such as Firefox, Google Chrome, Safari, Opera or IE 9.0+ for the best display.
The RCDdb database is freely available to the research community using the web link (http://chenyclab.com/RCDdb). Users are not required to register or login to access features in the database.
The RCDdb database is freely available to the research community using the web link (http://chenyclab.com/RCDdb). Users are not required to register or login to access features in the database.